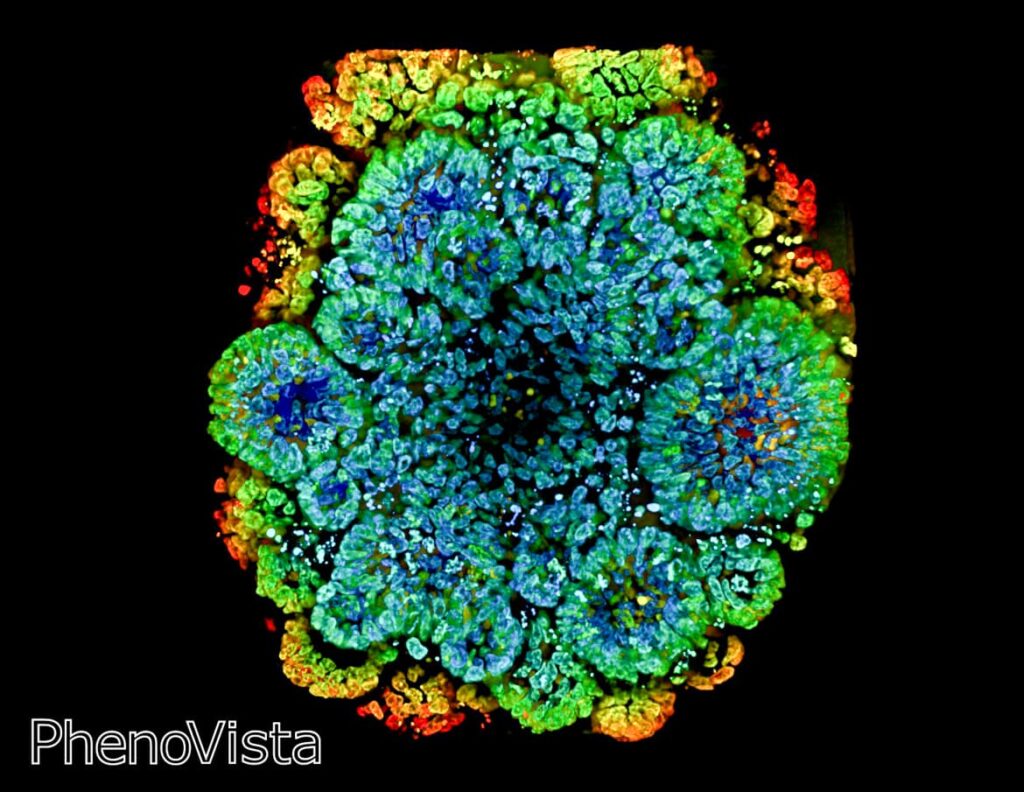
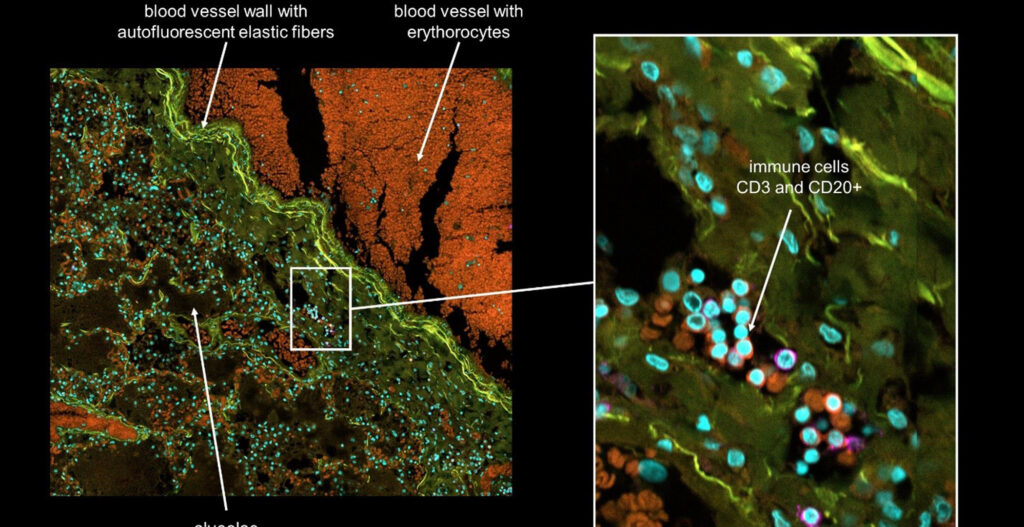
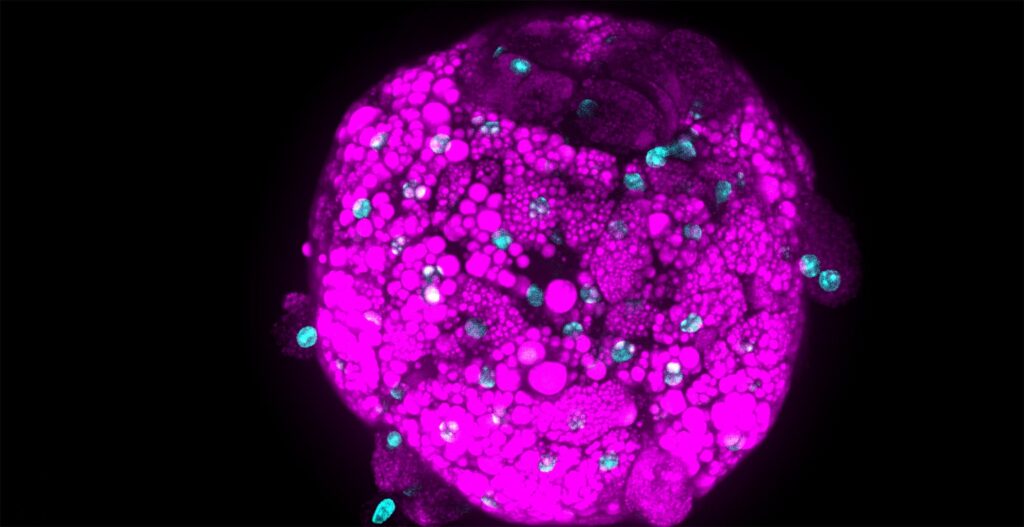
Introduction to PhenoVista Business
PhenoVista provides high-content, imaging-based research services on a contractual basis to pharmaceutical and biotechnology clients working across diverse biology/disease areas, including neurobiology, rare disease, immunology, oncology, animal health, and beyond. Clients choose to partner with us to access our core expertise in the design, development, execution, and analysis of high-content imaging assays, to deliver data derived from physiologically relevant, often complex 2D and 3D cell models aligned with their biology of interest.

What was challenging for your business and what was needed to resolve it?
Our client-project designs range from completely bespoke, development projects to straightforward onboarding and analysis of client-generated samples. We routinely design and develop imaging assay workflows using highly multiplexed markers, such as patient-derived organoids, cell painting, as well as working with mono- and co-cultured 2D and 3D cellular models built with, for instance, primary patient- and iPSC-derived cells. From these models, we generate diverse readouts at the cellular and subcellular levels – from whole-cell marker positivity to nuclear/organellar localization, and beyond. Some general, commonly requested assays include phenotypic cell painting, evaluation of mitochondrial or cellular health, uptake and localization of test articles, and detection and co-localization analysis of markers of interest. To accommodate this wide range of sample types and imaging applications, we built a suite of imaging instrumentation, including several CQ1 instruments, which offers a balance of ease-of-use, throughput, flexibility, and image/data quality.
How are the CQ1 systems used for your business?
We routinely use our CQ1 systems to capture data from, for instance, 2D co-culture assays generated using iPSC and primary cells, and cell painting studies, which require high-resolution imaging capabilities coupled with the flexibility to accurately image up to five fluorescent markers in this multiplexed format assay. The CQ1 is also our first choice for executing customer projects requiring high-resolution imaging data, particularly from the more complex cell models such as 3D co-cultures (including 3D bio-printed samples), 3D spheroids/organoids, micropattern plates, and more.
For example, we collaborated with Cellesce (https://cellesce.com/) to characterize cell-health marker responses of patient-derived, cancer-relevant organoids to standard-of-care oncology drugs, including the representative images and data highlighted below, using the CQ1 coupled with the CellPathfinder software (Figure 1).
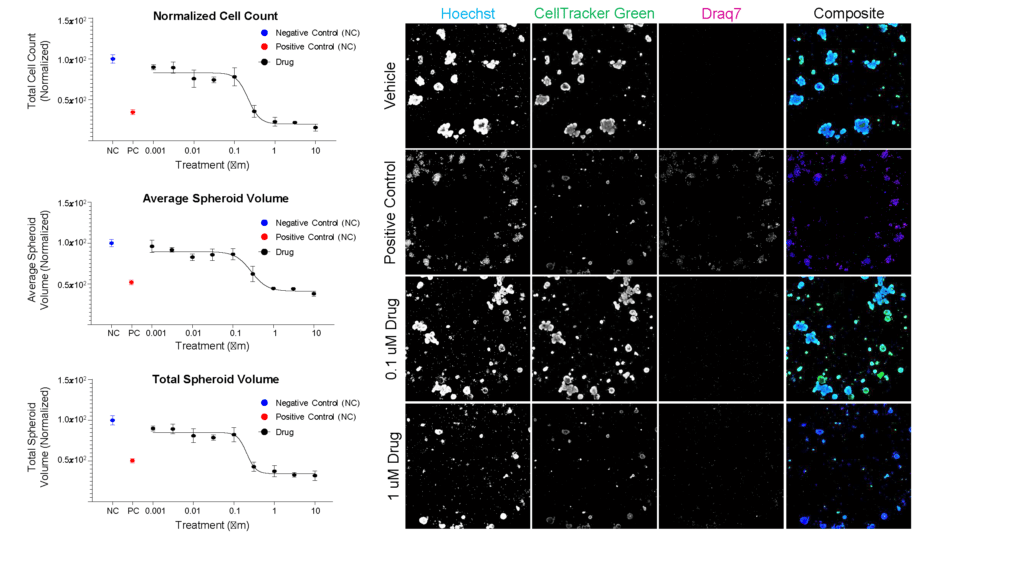
Ready-to-screen cancer organoids (Cellesce) were seeded in 384 well imaging plates, treated for five days with dose range of a standard-of-care cancer drug, then stained/fixed to detect nuclei (Hoechst), live cells (CellTracker Green), and dead cells (Draq7). The Yokogawa CQ1 imager was used to capture Z-stack images of these samples at 10X magnification. The CellPathfinder software was then used for image analysis, including quantitation of organoid volume, to assess dose-dependent impact of drug treatment.
Benefits of the CellVoyager CQ1 system
In addition to its excellent confocal image quality, the CQ1’s flexibility to accommodate analysis of highly multiplexed markers and optical sectioning (z-stack capture) capabilities make it an ideal platform for running detailed mode-of-action studies that require complex, high-resolution, 2D/3D analysis, as well as for higher-throughput cell painting projects. The CQ1’s overall ease-of-use and small physical footprint allow us to easily operate multiple instruments in parallel to increase throughput without compromising image/data quality.
Prospects for the future on developing drug discovery pipelines
As profiling of therapeutics in vivo/animal models becomes less common, we may see increased interest in developing and utilizing more complex, physiologically relevant in vitro human cell-based models, such as 3D organoids and organs-on-a-chip, to fill that translational gap. Additionally, we expect to see continued/growing interest in phenotypic profiling platforms, such as cell painting, to help classify the activities of orphaned drugs, drive repurposing of approved therapeutics, etc. That said, we anticipate that our CQ1 systems will help us to address the increased demand for these types of projects.
References
Bray, Mark-Anthony, et.al. Cell Painting, a high-content image-based assay for morphological profiling using multiplexed fluorescent dyes. Nature Protocols 11, pages1757–1774 (2016). (https://www.nature.com/articles/nprot.2016.105)