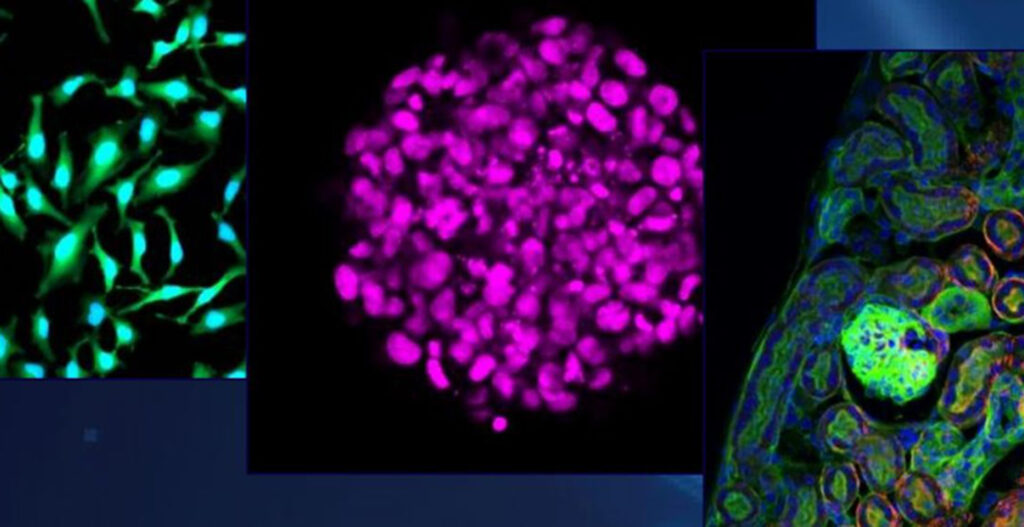
The first data (1) was published with SS2000 in Europe with our collaboration partner at the University of Surrey (Prof. Melanie Bailey’s lab).
What are Lipids?
Lipids are organic compounds consistent with nonpolar molecules, which are soluble only in nonpolar solvents (e.g., chloroform) and insoluble in water because water is a polar molecule. In the human body, these molecules can be synthesized in the liver.
Lipids are an essential part of a cell`s biomolecular pool and play important roles in a myriad of complex biological processes like:
- Supporting cells and aiding in essential functions
- Protecting nerve cells
- Helping the body absorb certain vitamins
- Helping produce hormones, including estrogen, testosterone, and cortisol
- Energy storage (in the form of fat)
- The structural component of the cells
- Signaling functions
Changes in lipid composition are significant in fundamental biological processes such as growth, development, differentiation, and aging and are also important in disease states such as cancer. Being able to detect and quantify lipid composition in single cells would provide enhanced data on such cell states and reveal new information on the cellular role(s) of lipid heterogeneity.
For example, cancer cells show a wide variety of lipid profiles at the single-cell level. Cancer cells are highly heterogeneous and very diverse in their phenotype which makes them hard to extinguish, but the analysis of single cells and the characterization of the heterogeneity could provide new treatment strategies.
Lipidomics: Analysis of Lipid Profiles
Lipidomics is a newly emerged discipline that studies cellular lipids on a large scale based on analytical chemistry principles and technological tools, particularly mass spectrometry (MS). Recently, techniques have greatly advanced, and novel applications of lipidomics in the biomedical sciences have emerged.
MS-based techniques for the analysis of lipids are different in the absence or presence of liquid chromatography (LC) prior to mass spectrometric analysis. They can also be different in their analytical coverage (e.g., “targeted” vs “global” analysis).
LC is a separation technique for lipids in which the mobile phase is a liquid that can be carried out either in a column or a plane.
Challenges for Analysis of Lipid Profiles
One of the challenges is the low abundance of analytes present in one cell. There are new methods emerging on the marked dealing with such challenges. Another challenge is the disturbance of the natural state of cells by current instruments like microfluidic systems or machines that are droplet-based. The cells lose their spatial context by being detached from the surface they are cultivated on. Also, minor changes to the environment, which stress the cells, can change their lipid profile.
The sensitivity also plays a pivotal role in single-cell lipidomic studies and is another challenge that needs to be addressed.
Single-Cell Lipidomics Performance after SS2000 Single-Cell Sampling
For the first time, researchers at Surrey University could, with the help of SS2000, combine live cell imaging of pancreatic cancer cells with single-cell lipidomics. The results show an increase in the lipid droplet size (figure C) which was detected based on fluorescence microscopy and found to correlate with the higher abundance of neutral DG (Diglyceride) and TG (Triglyceride) lipids after induced oxidative stress (figure D).
This novel methodology has demonstrated unique capabilities. These were otherwise inaccessible to mass spectrometry imaging because living, single cells were sampled, and fluorescent live cell imaging was used on the same cell culture from which cells were sampled.
To demonstrate the capability of the developed lipidomics method for biological applications, the researchers sampled living single cells from two different cell cultures (PANC-1 and THP-1) and analyzed their lipid profiles. The results showed that the two groups separated significantly (figure A). This means that different cell types can be distinguished based on their lipidome.
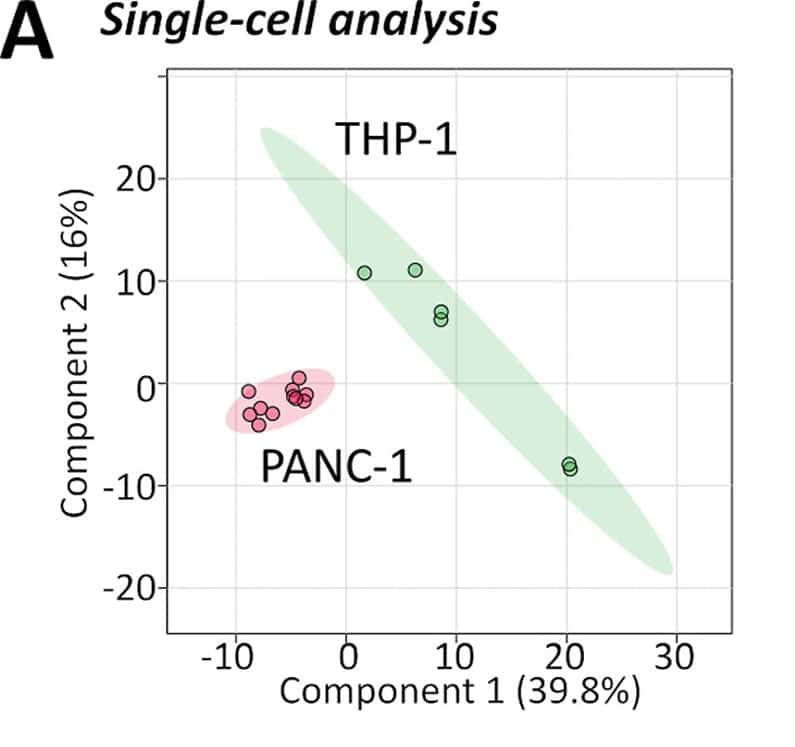
The success of this methodology has been demonstrated in its ability to distinguish different cell types based on their lipidome (figure A) and make single-cell lipidomic observations consistent with previous observations in the literature based on cell extract analysis (figure B-D).

Why study Cancer by Single-Cell Lipidomics?
Lipids play many key roles in all basic processes essential for tumor development:
1. Lipids affect cell growth and metabolism, which are essential for rapidly proliferating cancer cells as the major building blocks for lipid biosynthesis and remodeling:
- Lipids represent the major structural components of cellular membranes (cholesterol, phospholipids, and sphingolipids)
- Lipids serve as the energy storage depot (triglycerides)
- Lipids are involved in energy metabolism and ATP production (acyl CoA and acylcarnitine)
2. Lipids are important for cell signaling:
- Lipids function as second messengers and as hormones in cancer cells to promote cell proliferation, survival, and migration (bioactive lipids, such as lysophospholipids)
Lipids are involved in significant cell signaling processes in chemotherapy and radiotherapy for human cancers.
We wish Dr. Melanie Bailey, Dr. Johanna von Gerichten, and Kyle D. G. Saunders a lot of success with their further research.
Learn more about our collaboration project SEISMIC with the University of Surrey!