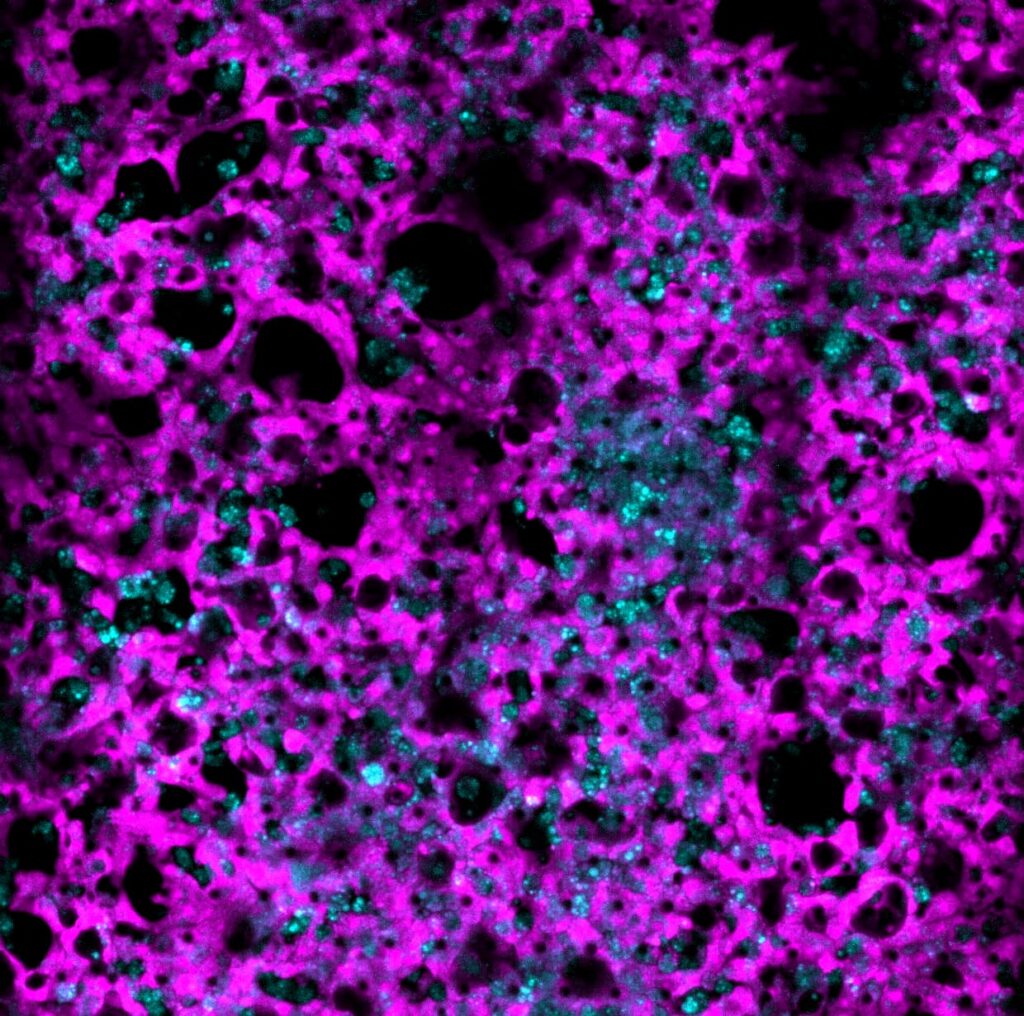
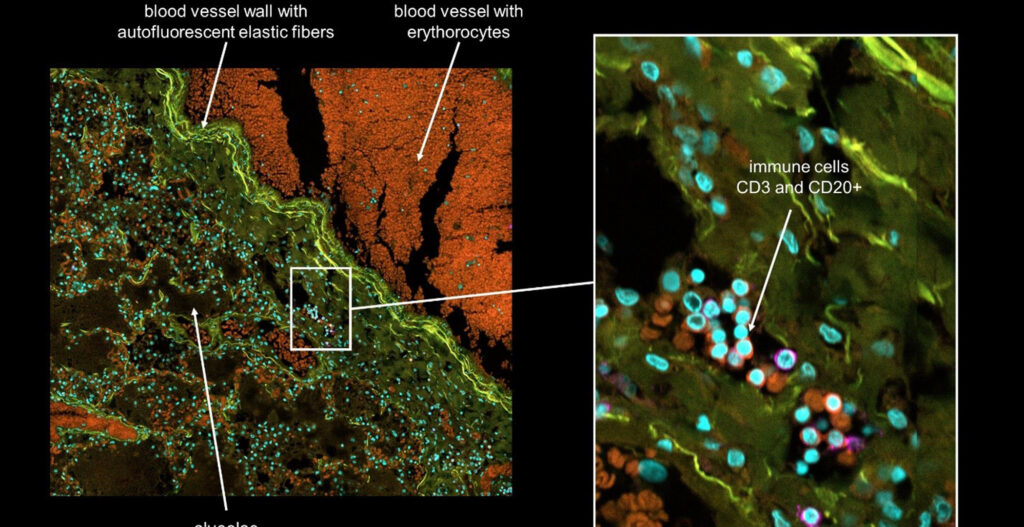
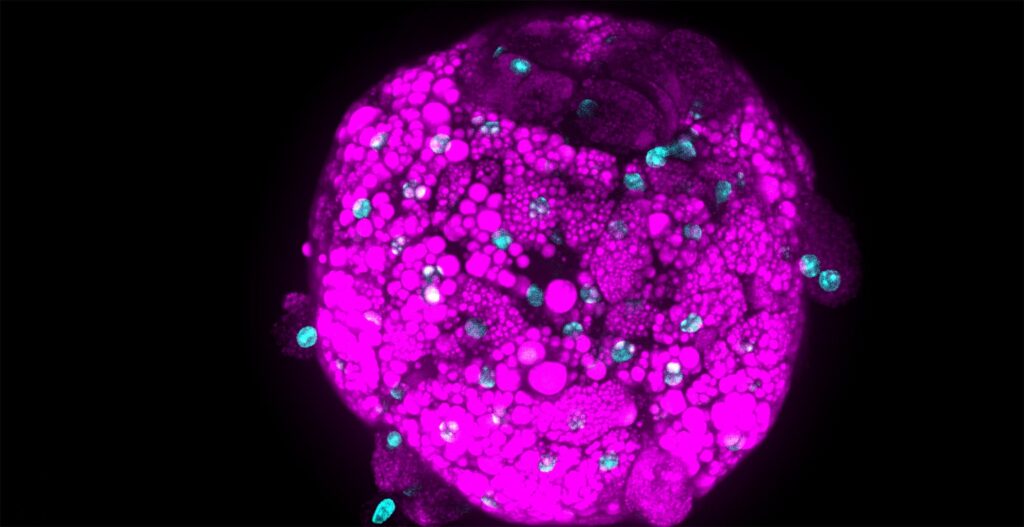
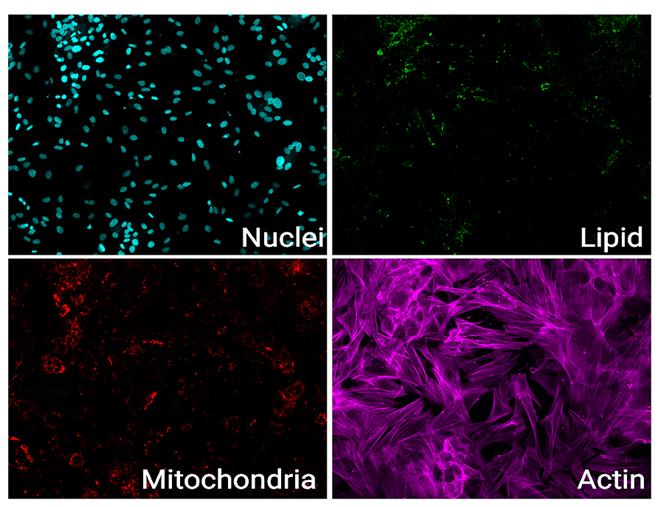
As immortalized cell lines and animal testing are continuously demonstrated as poor models for human biology, there is a big movement towards conducting assays in 3D organoid systems. As powerful as organoids are, there are limitations in the scalability of their culture and therefore implementation in high-throughput drug screening assays. Sexton lab at the University of Michigan, US focuses on developing bioassays evaluating organoid and other complex cell systems through a multi-omics approach followed by an adaptation of assays into high-throughput imaging-capable platforms for rapid drug screening. Their methods require the rapid acquisition of high-quality confocal images in 3-dimensional space with certain assays also requiring live-cell imaging.
Introduction to your research
Drug-induced liver injury (DILI) is a hindrance to both drug development and patient well-being. DILI is not well understood as no current models, both in vitro and in vivo, are consistent with representing the human liver function. Patient genetic diversity also plays a key role in DILI, especially in idiosyncratic DILI. We are currently developing a panel of iPSC-derived human liver organoids from a genetically diverse pool of patients who have suffered from idiosyncratic DILI in an attempt to model hepatotoxicity in a laboratory setting. The profiling of the genomic, transcriptomic, and phenotypic identity of each patient-derived organoid line is ongoing.

What is challenging and a bottleneck for your research?
As no definitive single endpoint exists in measuring DILI, nor does DILI manifest the same across drugs and patient lines, we are reliant on multivariate phenotypic endpoints to visualize and measure DILI. As adaptive immunity is thought to be a key component of idiosyncratic DILI, and the liver is a complex multicellular organ, advanced multicellular culture systems involving parenchymal hepatocytes, non-parenchymal cells, and immune cells are necessary.
High-content imaging/analysis is viable in this case as a relatively low-cost and scalable system to immediately visualize cells, delineate cell types, obtain cell-level measurements, and incorporate multivariate analysis in describing DILI all from a single assay. However, rapid acquisition of the necessary image data in a time-efficient manner is a bottleneck, especially in the context of live cell imaging.
Why do you use the CellVoyager™ CQ1 and CV8000 in your research?
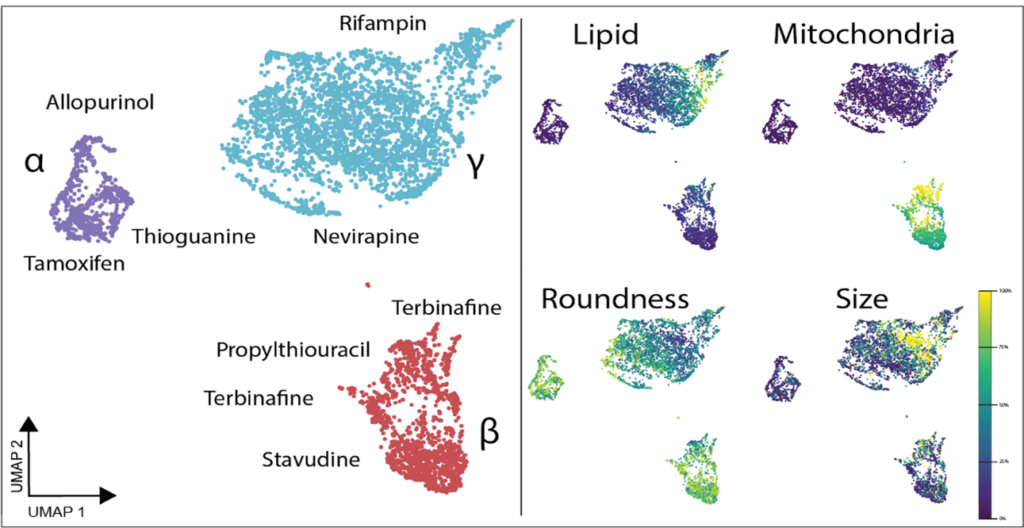
Both the Yokogawa CellVoyager CQ1 and CV8000 systems have been crucial in our research. The CQ1 serves as a straightforward, easy-to-use, but feature-heavy platform that has been amenable to different assays. We have successfully optimized imaging for standard cell culture plates of all sizes and non-standard organ-on-chip platforms. For example, we are now using image-based profiling of cells to capture morphological perturbations caused by herbal and dietary supplements (HDS) that have been implicated in DILI. With the cell-level feature matrix, we are able to identify HDS products that don’t necessarily kill cells, but cause a large phenotypic shift as measured with distance score compared to control (Figure 1).
As both CQ1 and CV8000 have full cell culture capabilities, we have also been able to leverage live-cell imaging assays. As many forms of DILI involve immune responses we have employed PBMC-organoid co-culture assays to visualize PBMC recruitment and attack of organoids. These assays are done in 384-plates and run for over 24 hours with each organoid imaged every 10 minutes. This means that each well is imaged prior to the next time point. With follow-up analysis by the built-in CellPathfinder software, we have been able to capture interesting and informative results on PBMC movement and dynamics which we are applying towards studying idiosyncratic DILI (Movie 1).
Prospects for the Future
Going forward, we are adding complexity to our models with novel chip culture systems and co-culturing with additional cell types. Both the CQ1 and CV8000 with play important roles in capturing images to not only obtain single-cell resolution but also for delineating cell type, spatial information, and in the case of chip culture, various compartments.
Lab Website
Sexton Laboratory – Diabetes and obesity research at the University of Michigan