Dr. Mancini's lab focuses on single-cell analysis specifically transcription by steroid receptors primarily estrogen and androgen receptors.
Please tell us about your research.
I started my lab at the Baylor College Medicine in 1995 after completing postdoctoral research in 1995. Originally, I spent a lot of time utilizing electron microscopy, but fluorescent microscopy was introduced and became the dominant method of imaging. Increasingly bigger, faster, and stronger microscopes obviated the need for electron microscopy in a lot of situations, but not all.
We focus on single-cell analysis specifically transcription by steroid receptors primarily estrogen and androgen receptors. The department is well known in the steroid receptor and nuclear receptor field. I saw an opportunity to bring imaging to a field that was largely biochemistry oriented. As molecular biology took off in the 1980’s, cloning became more prevalent. The tools to perform imaging in that same context are a user’s ability to clone fluorescent proteins to make fusions and follow them in the microscope, and that has been the main source of what we do.
In the last 15 years has the speed of high-content imaging improved? Tremendously, originally we used cover slips and there are only so many cover slips that can be done. When fluorescence microscopy and microtiter plates were introduced maybe 20 years ago, I signed up knowing the technology would be impactful in our research. Once we made the connection with automated high throughput we never looked back.

Left to Right: Hannah Johnson, Fabio Stossi,
Michael Mancini
How has high-content imaging helped progress your research?
Speed and quantitation, we designed single-cell oriented tissue culture models that were amenable to much more than usual. Yes, we could attain a nice picture of it and see the receptors inside the nucleus or cytoplasm, but we could not see all the mechanistic parts we wanted to see. We could not see DNA binding, transcription, but we saw where the receptor was.
I have made many permutations of a cell line where we engineered a visible transcription locus. That allowed us to do more measurements that were mechanistic. We were able to measure how much protein is the cell, how many proteins are in the nucleus versus the cytoplasm. We can visualize DNA binding because we integrated and reported genes enough to be able to watch the locus expand and contract based upon antagonism or agonism, hormones or antihormones, and we can visualize transcription. All that in one experiment
What role does the Yokogawa CellVoyager CV8000 High-Content Screening System play in your research?
This has been a somewhat miraculous jump to faster results and more capabilities. More channels simultaneously, the speed is incredible, and the quality of the image is so much better than our previous high-content imaging system. We use our old system as a backup machine, but I don’t think anyone wants to take pictures with it anymore as long as we have the CellVoyager CV8000 High-Content Screening System.
What is the key to making full use of the CV8000's capabilities?
Design your experiments to take advantage of what the device can do. The cell painting is a new avenue that the CV8000 is built for. The CV8000 also allows the user to utilize 4 or 5 channels simultaneously at a speed and resolution so much better than what we have had. All good and fantastic.

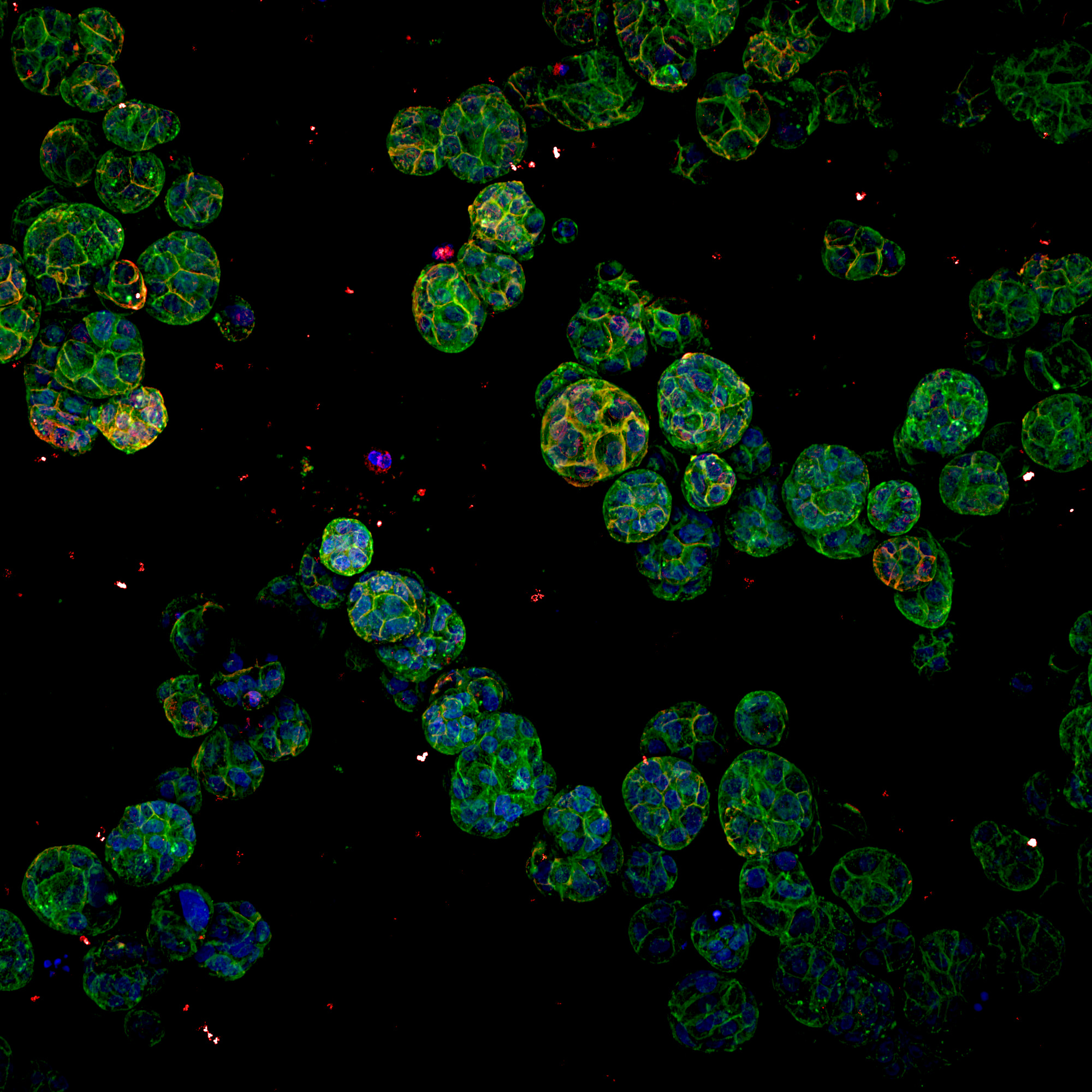
MCF10A (normal breast cells), grown in a 3D media. They were grown for 10 days in a 384 well plate. Staining, 4%PFA for 20 mins at room temperature, then Phalloidin added in the 488 channel, Wheat Germ Agglutinin (WGA) in the 555 channel. Labeled with DAPI for the nuclei and left the dye in for 3 days to get even staining with 0.1% TX-100. The image is a max intensity projection of the whole z stack, which was roughly 60 microns total, taken using the 20XWI objective.
What are the future plans for your research?
A combination of mechanistic basic science, how does this happen. We had an important paper, with Fabio Stossi as the lead author, visualizing genes that are turning on innative cells. No cellular engineering, it is done through mRNA fish, you learn not every cell fires the same number of alleles. On the cell line we were using there were four target gene alleles for the estrogen receptor and depending on how long the hormones were there you would see zero, one, two, three, or four alleles. Bringing in the statistics and the ability to take thousands of pictures you realize there is an order and something mechanistically controlling how many endogenous alleles are firing from the receptor. This turned out to be a short, but effective library screen with known inhibitors of epigenetic mechanisms. We learned about the epigenetic control of the number of alleles that are firing.
At the same time due to the throughput, ability, and power of high-content screening we have been involved in receptor biology connected to environmental issues. Our team has been a part of a fun Texas A&M research program for the last 4 ½ years. Their team has been focusing on contaminants present in the Houston Shipping Channel. We receive samples from Texas A&M, place them into our system, and see if there were any known compounds of interest in the sediment samples. We have some libraries with dozens of different compounds. We can rapidly evaluate whether there are any compounds of interest present in a matter of minutes using the microscope, so it does have a commercial industrial component as well.
What do you do when you’re not in the lab?
Think about the lab. In between that I try to find time to take advantage of photography. In fact, the pandemic has allowed me to buy an electronic keyboard which I haven’t played since I was a teenager. So, it has been fun learning how to play the keyboard again. The way I learned as a kid was always sheet music, which was always assigned to me, so I never had a choice of what to play. As an adult I play what I feel like playing, which could be freestyling on the keyboard or guitar.
Michael Mancini Ph.D.
https://www.bcm.edu/people-search/michael-mancini-26042
Professor, Baylor College of Medicine
Department of Molecular Cell Biology and Pharmacology Cross appointed at Texas A&M’s Institute for Biosciences and Technology IET
Interview date: 2022
Our Social Medias
We post our information to the following SNSs. Please follow us.
| Follow us | Share our reference | |
| @Yokogawa_LS | Share on Twitter | |
| Yokogawa Life Science | Share on Facebook | |
| Yokogawa Life Science | Share on LinkedIn |
Yokogawa's Official Social Media Account List
Related Products & Solutions
-
CV8000 High-Throughput System
Simplify and accelerate workflow processes with the CV8000.
-
Life Science
- Yokogawa life science products
- High content analysis systems
- Dual spinning disk confocal technologies
- High-speed, high-resolution live-cell imaging
- Enabling cutting-edge research

